Release Information
Current DbStRiPs release: January, 2021
PDB version: The DbStRiPs database is constructed by considering all protein chains
reported in the Protein Data Bank release of June 3, 2019. It includes 502,599 protein chains
reported in PDB format, 50,419 chains of large compexes and 2,317 chains in model structures.
UniProt version: 2019_11 (November, 2019)
Browse page
The browse page represents the classification scheme of the DbStRiPs database.
Clicking a repeat family leads to the 'Repeat family page'.
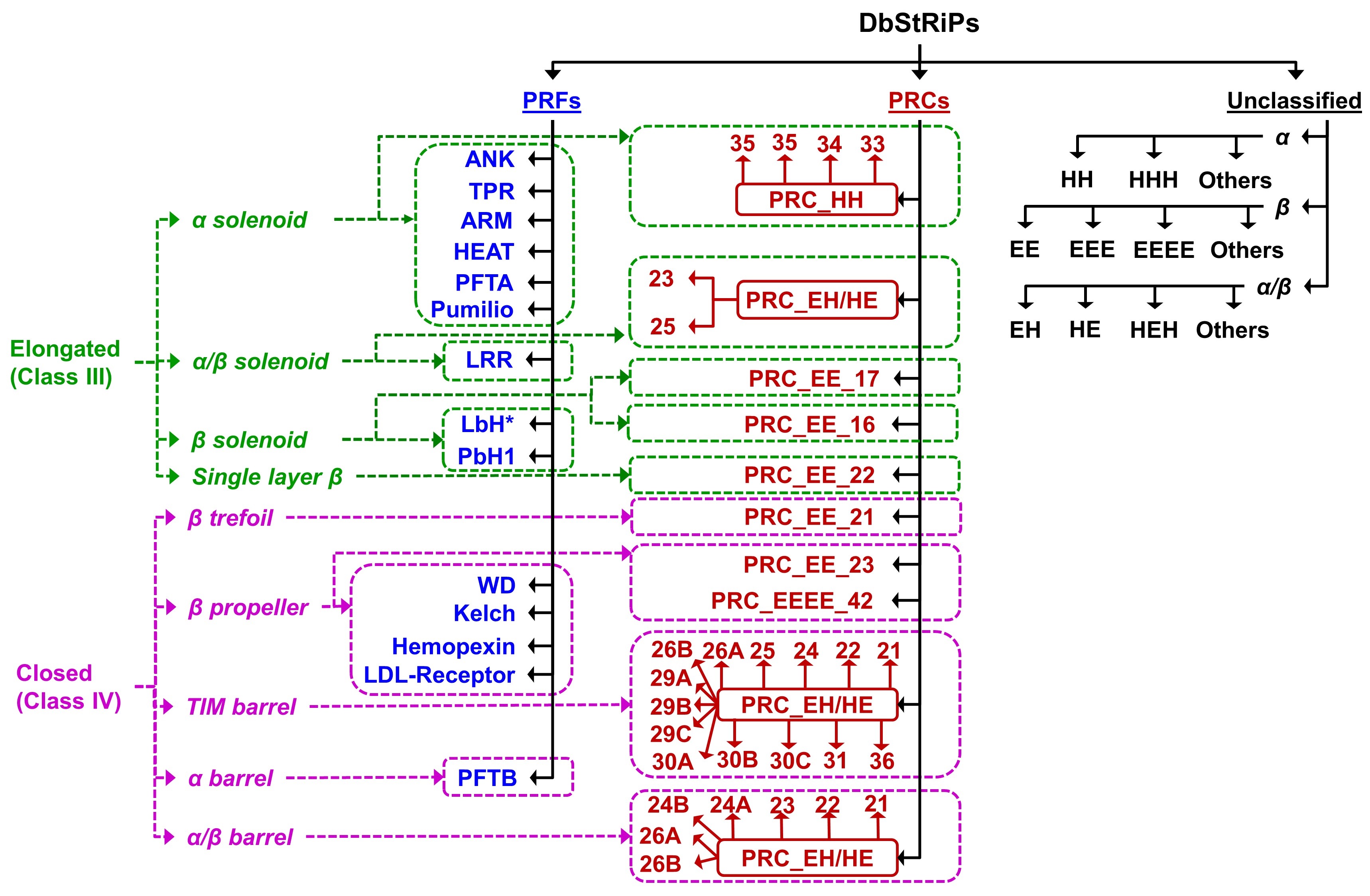
Classification schema of DbStRiPs
* marked LbH is novel repeat family. The families starting with name "PRC" are Protein Repeat Clusters
obtained from MCL clustering followed by Clique analyses, and classified based on CATH architecture.
The name of these clusters are given as "PRC_<secondary structure>_<architecture>_<motif length>".

Classification schema of DbStRiPs
Repeat family page
The repeat family page provides details about a particular repeat family. The details include:
1) Description: A short description of the repeat family derived form InterPro description
giving details of the repeat architecture, abundance and function. The number of chains
predicted with the repeat family is also given.
2) Links to other databases: Links to the following databases (whenever available) are provided-
InterPro, PROSITE, Pfam, PRINTS, SMART.
3) Table: Table giving details of all the PDB chains belonging to the repeat family. The table includes:
i) PDB Id
ii) Chain
iii) UniProt Id
iv) Name: Represents the UniProt name
v) Organism: Extracted from UniProt
vi) Repeat annotation: DbStRiPs repeat annotation
vii) Copy number: Number of repeat copies
viii) Score: Overall score of prediction
ix) View: Redirects to corresponding PDB page
The table can be sorted based on PDB Id, UniProt Id, Copy number and Score.
Download: The table with all the above mentioned fields can be downloaded in text (tab separated) and
CSV formats.
PDB page
The PDB chain page shows the details for each PDB chain. The details include:
1) PDB details: The details extracted from the Protein Data Bank are shown. This includes:
i) PDB id
ii) Chain id
iii) Experimental method used to resolve the structure
iv) Resolution of the structure
v) Structure title of the PDB file
2) UniProt details: The details extracted from the UniProt entry for the protein are shown. This includes:
i) UniProt id
ii) UniProt name
iii) Organism
iv) Disease association: Disease association is extracted from UniProt and linked to MIM id of OMIM database
3) 3D structure: Browser independent JSmol applet is used to show an interactive three dimensional
structure of the PDB chain. The secondary structure assignment in JSmol is taken from author's secondary structure
assignment given in PDB file, which may not match with the STRIDE/DSSP assignment used by the backend algorithm PRIGSA.
The image of the protein structure can be saved in PNG format using 'Save Structure' button.
4) Repeat Type/Periodicity: The repeat types predicted in a PDB chain are shown in the form of a
radio button. On clicking a radio button, the predicted repeat boundaries for the repeat type are
shown. The repeat copies are color coded on the JSmol applet so that the user can correlate the
copies on the 3D structure.
5) Number of copies: Shows the number of repeat units of the selected repeat type.
6) Average Score: Average score of all copies of a particular type.
7) Repeat copies: Shows the details of repeat copies.
i) Repeat boundary annotation: Start-end boundary of repeat copy.
ii) Secondary structure: Secondary structure assignment by STRIDE algorithm is shown for the repeat copy.
iii) Score: Indicates the confidence of prediction of the repeat unit. Higher values (closer to 1.00) indicate
more confident repeat unit prediction.
8) Multiple Sequence Alignment: MSA of the repeat copies can be carried out using Clustal Omega by clicking
on the button "Multiple Sequence Alignment". A new window will open showing the MSA of all copies with amino acids colored
according to the physico-chemical properties. The MSA can be downloaded in text format.
Search page
The menu bar on each page has a search option. The search can be performed by three ways:
1) Search by PDB id: On entering a valid PDB Id, all protein chains of the PDB record are shown.
2) Search by UniProt id: On entering a valid UniProt Id, all the PDB chains corresponding the
UniProt entry are shown.
The search result comprises of:
i) PDB id
ii) Chian
iii) UniProt id
iv) PDB length: The UniProt coordinates contained in the PDB entry.
v) View button: Redirects to PDB page.
3) Search by Keyword: On entering a keyord of length 3-20, the keyword is searched in repeat family names,
protein names (UniProt name) and PDB titles. In case of a repeat family, the family is shown in the result.
For keyword search in protein name and PDB titles, the search result will show all repeat chains having the keyword.
Note: the keyword search results in UniProt name or PDB title may contain some repeats missed by the KPRF module,
because it did not meet the stringent criteria, however, it might have been detected by the de novo module as a
repeat protein and may belong to PRCs or URPs.
Methods | |
Chakrabarty B, Parekh N: PRIGSA2: Improved version of protein repeat identification by graph spectral analysis. J Biosci 2020, 45(1):95.link
|
Chakrabarty B, Parekh N: Identifying tandem Ankyrin repeats in protein structures. BMC Bioinformatics 2014, 15:6599.link

