
 |
 |
 |
The main purpose of this database is to establish a correlation between occurrences of different base pairs in RNA structures in a particular geometry and their stability. We know that, not only the complex structures of RNAs but their structural dynamics determine their complex functionalities. Although, quantum mechanical (QM) calculations help to understand intrinsic stability (not considering the actual crystal context) of different base pairs, but understanding the intrinsic stability and ground state optimized geometry outside the crystal context can explain how structural environment can effect stability and local geometry of a particular base pair in experimental structures. This understanding is important for studying dynamics of larger RNA molecules. In this database we have collated all the important and available QM calculation data including interaction energy and its components for all possible (both observed and modeled) base pairing geometries. In our group an extensive study on base pairs, their geometries in crystal and ground state optimized structures and their intrinsic interaction energy has been carried out previously. There are other groups who are also involved in studying RNA base pairs using QM methods. Here we have arranged all those data systematically so that it can help both computational biologist who are studying RNA structures and dynamics and experimental biologist developing nucleic acid based nano-devices. This database also can help in modeling new RNA structures by providing geometry and stability related data of different base pairs. The following sections will help you navigate through the database.
Go through the Home page. Figure 1 shows a screenshot of RNABP COGEST home. This provides a brief overview of the database. The options marked with green coloured numeric identifiers are described in detail here.
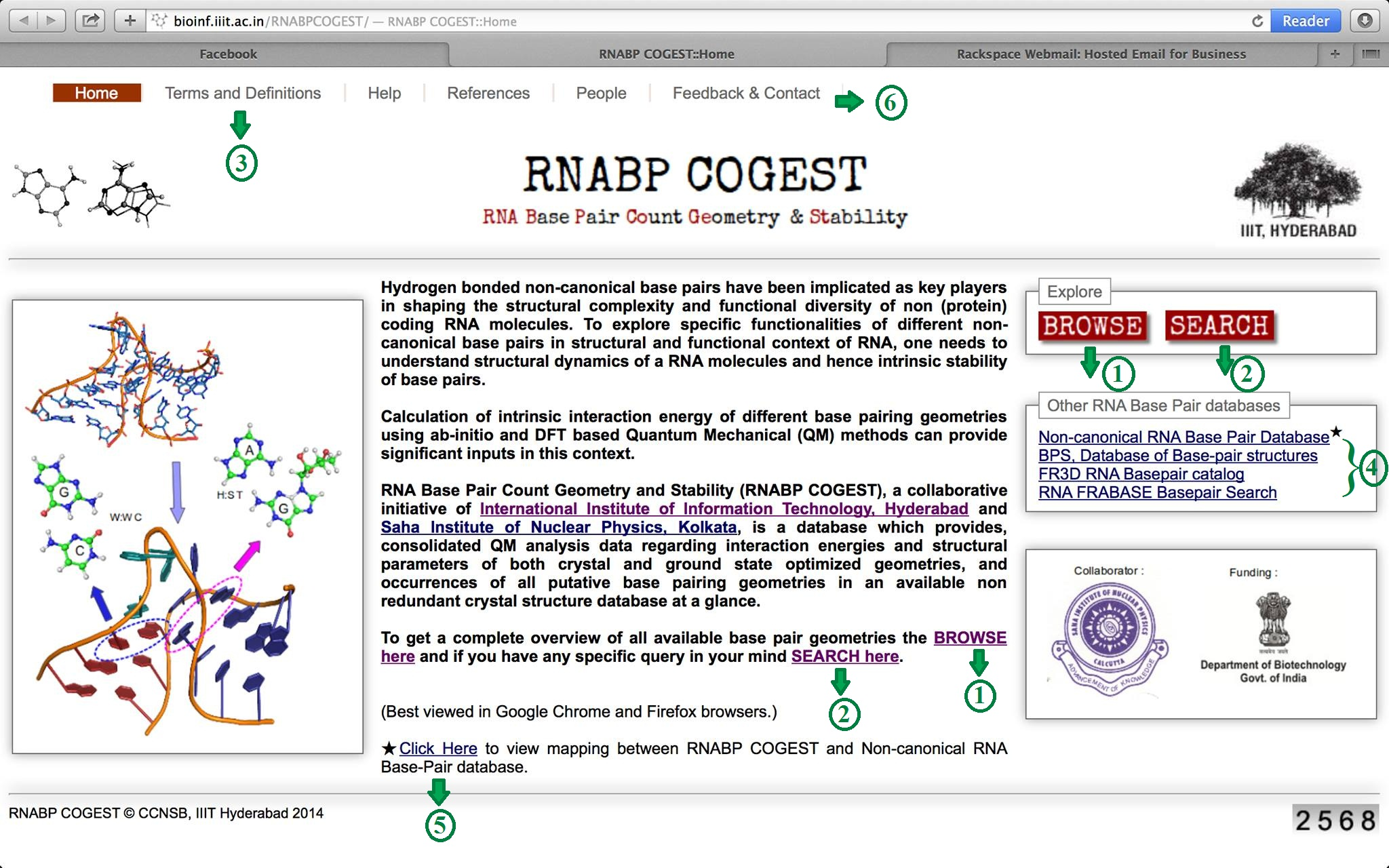
Identifier 1 Identifier 2 Identifier 3 Identifier 4 Identifier 5 Identifier 6
This will redirect here. It is a matrix representation of all possible base pair combinations. Base pairs are characterized by identity, interacting edges and respective glycosidic bond orientation of pairing bases. Based on first two criteria 144 base pairing combinations are possible and hence, 144 small boxes are there in the matrix representing each combination. In all the boxes there are radio buttons, by selecting anyone and pressing search button below the matrix you can get a overall summary about that particular base pairing geometry.
For some geometry due to absence of suitable hydrogen bond donor and acceptor atoms in interacting edges, those geometries are forbidden completely. Radio buttons are not present in those boxes as shown in Figure 2.
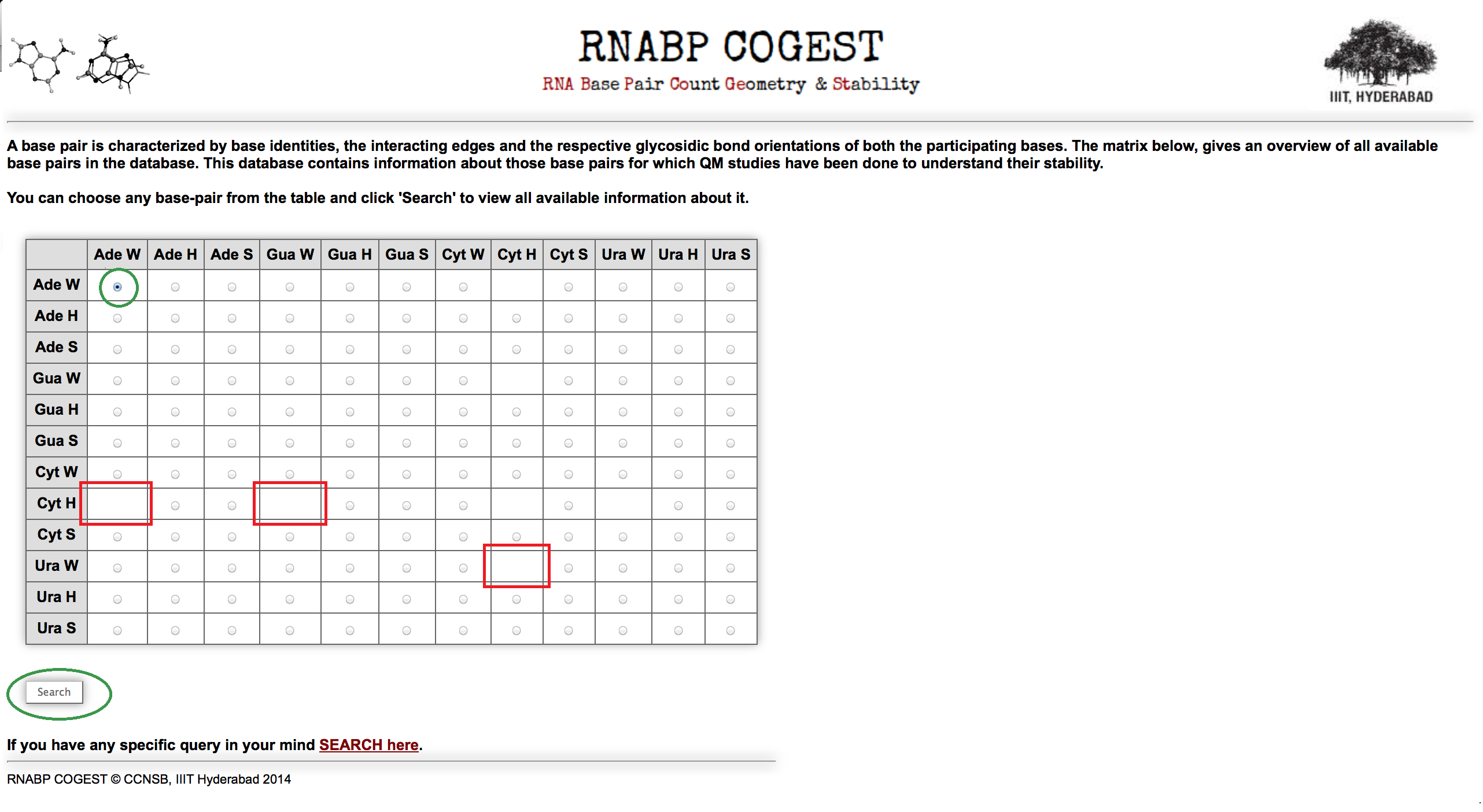
Suppose you have chosen Ade W:Ade W combination from the matrix (highlighted in green circle in Figure 2) and make a search for it. It will redirect you to an overall base pair information page.
In each combination depending on respective glycosidic bond orientation two distinct configurations (cis and trans) are possible. Again, multiple examples of each base pairing geometries are there based on protonation, hydrogen bonding mode (conventional or amino acceptor), mutual positioning of two participating bases in crystal geometry (adjacent or distant), and even on model building criteria considered during optimization (sugar moiety replaced by methyl, hydrogen etc.). Figure 3 represents the result of the previous search for Ade W:Ade W base pairing family.
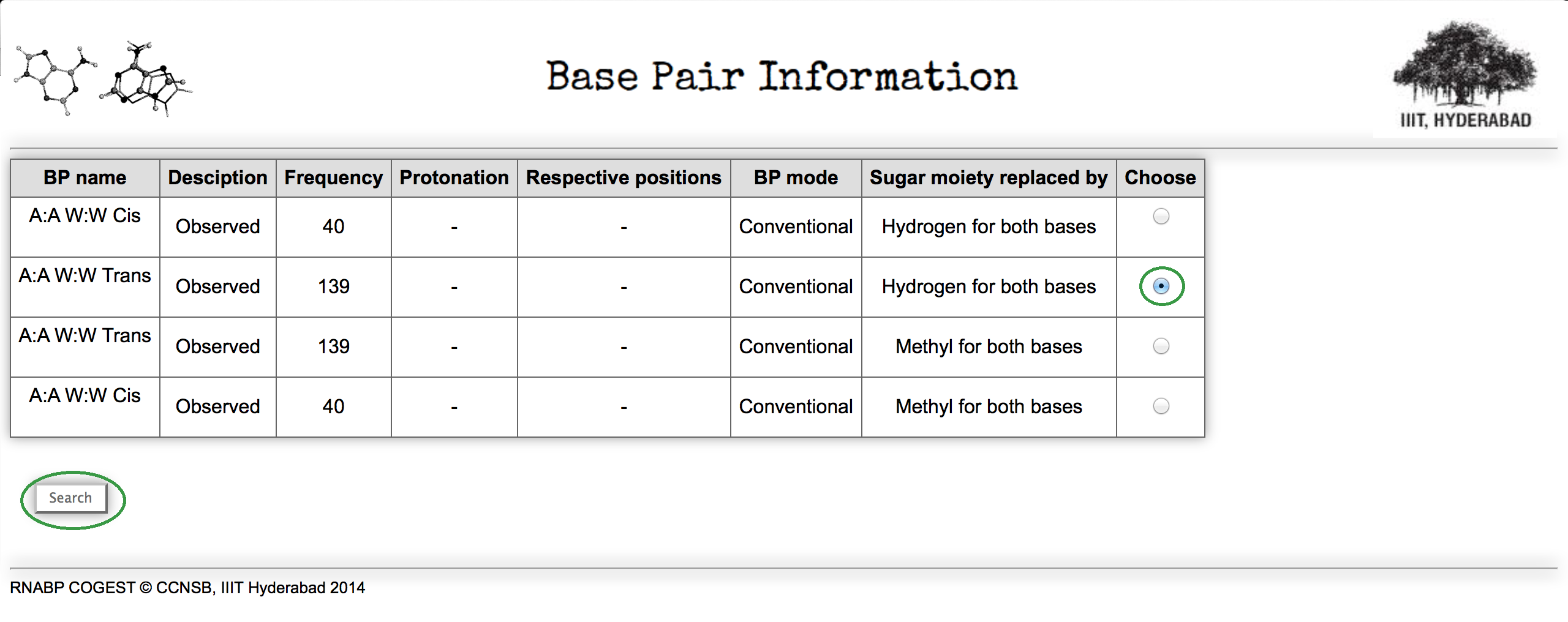
The table is more or less self explanatory. To understand the definition of BP mode Click Here.
Now if you choose any one from the list and click the search button, it will redirect you to a detailed information page. Suppose, we have chosen A:A W:W Trans base pair from the table for which sugar moiety of both the bases are replaced by simple hydrogen atoms.
This is a long information page providing all available information in the database about that particular chosen base pair (A:A W:W Trans). Each table is considered separately and explained here.
Table 1 marked in Figure 4 consists of general information of that base pair family and how it has been modeled to carry out a ground state optimization.
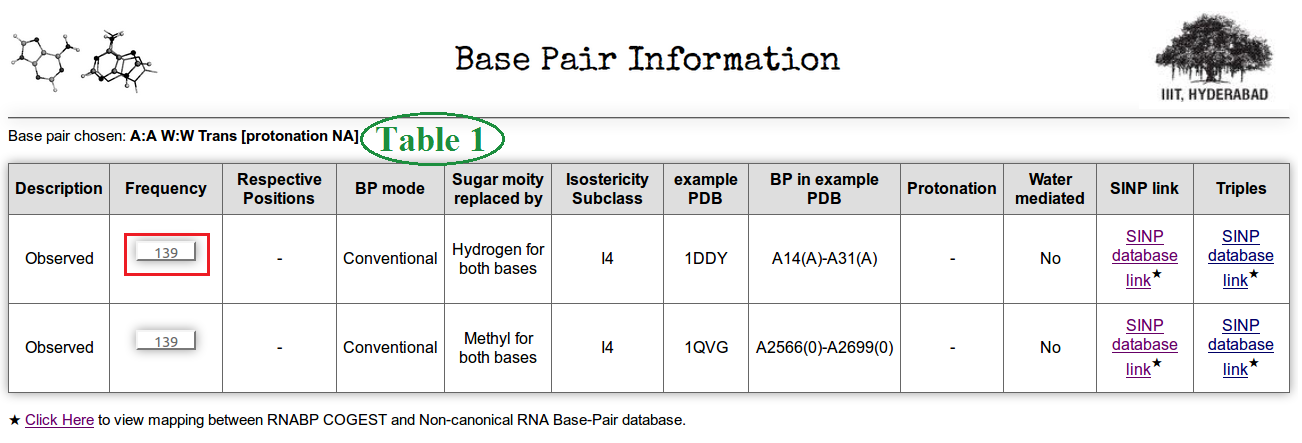
Column 2 of the Table 1 displays frequency of A:A W:W Trans base pairs in a non redundant dataset of RNA crystal structure obtained from HD-RNAS database (Ray et al., Front Genet. 2012). The selected non-redundant dataset contains a total of 167 pdb files of different types, which have resolution better than 3.5 Angstrom and the chain length cutoff is greater than 30 nucleotides. Clicking on to this button (highlighted in red box) user can obtain all the occurrences of the base pairs with information of pdb_id, pdb chain name and nucleotide numbers in that particular pdb file. This page also provides information of the basic six base pair parameters of that particular instance. Figure 5 shows a screen shot of that page.
There are instances of some base pairs, which are designated as observed but the frequency value is 0. This is because of two reasons mainly. First, the base pair is not very much frequently observed in nature and not present in the non-redundant data set, that we have considered for frequency calculation. Second, we have used BPFIND tool to detect occurrences of different base pairs in a non redundant crystal structure data set. BPFIND detects base pairs, which are stabilized by at least 2 good hydrogen bonds. So, the base pair with single hydrogen bond, and with not a very good geometry in terms of donor acceptor distances and planarity, are not detected by BPFIND tool.
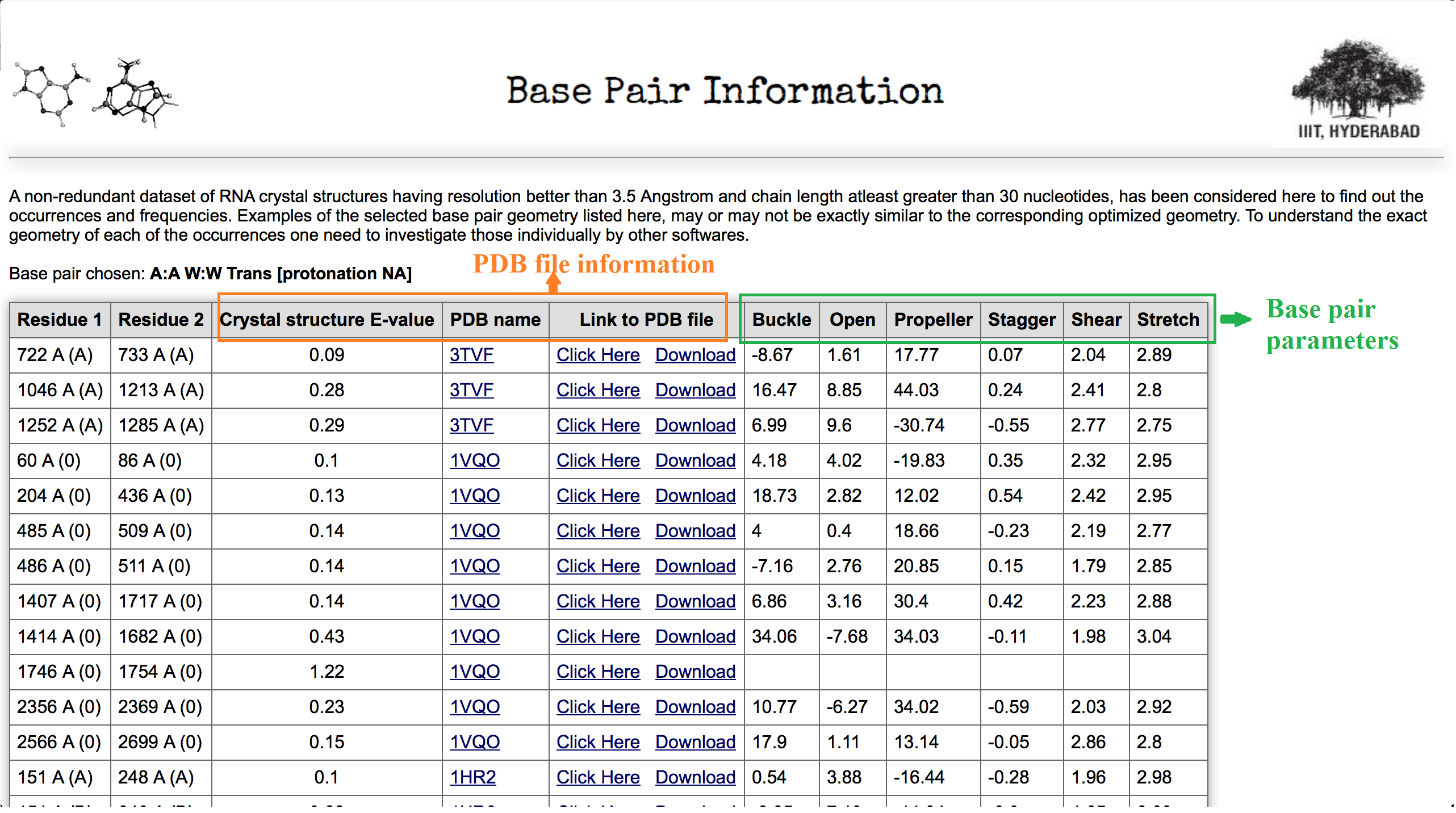
In the Figure 5 first two column of the table explains interacting residue information chain ids are within parenthesis. Parameters and pdb information are highlighted in Figure 5. Column 7 and 8 in Table 1 give information about base pairs taken for optimization and interaction energy calculation.
To understand BP mode and Isostericity subclass please Click Here.
Table 2 and Table 3 marked in Figure 6 provides information about base pair parameters optimized in different methods (where available). It can help you to compare different optimized geometry of same base pair. In Table 3 you can see the images and download coordinates of corresponding figures to visualize and edit in other visualization tools.
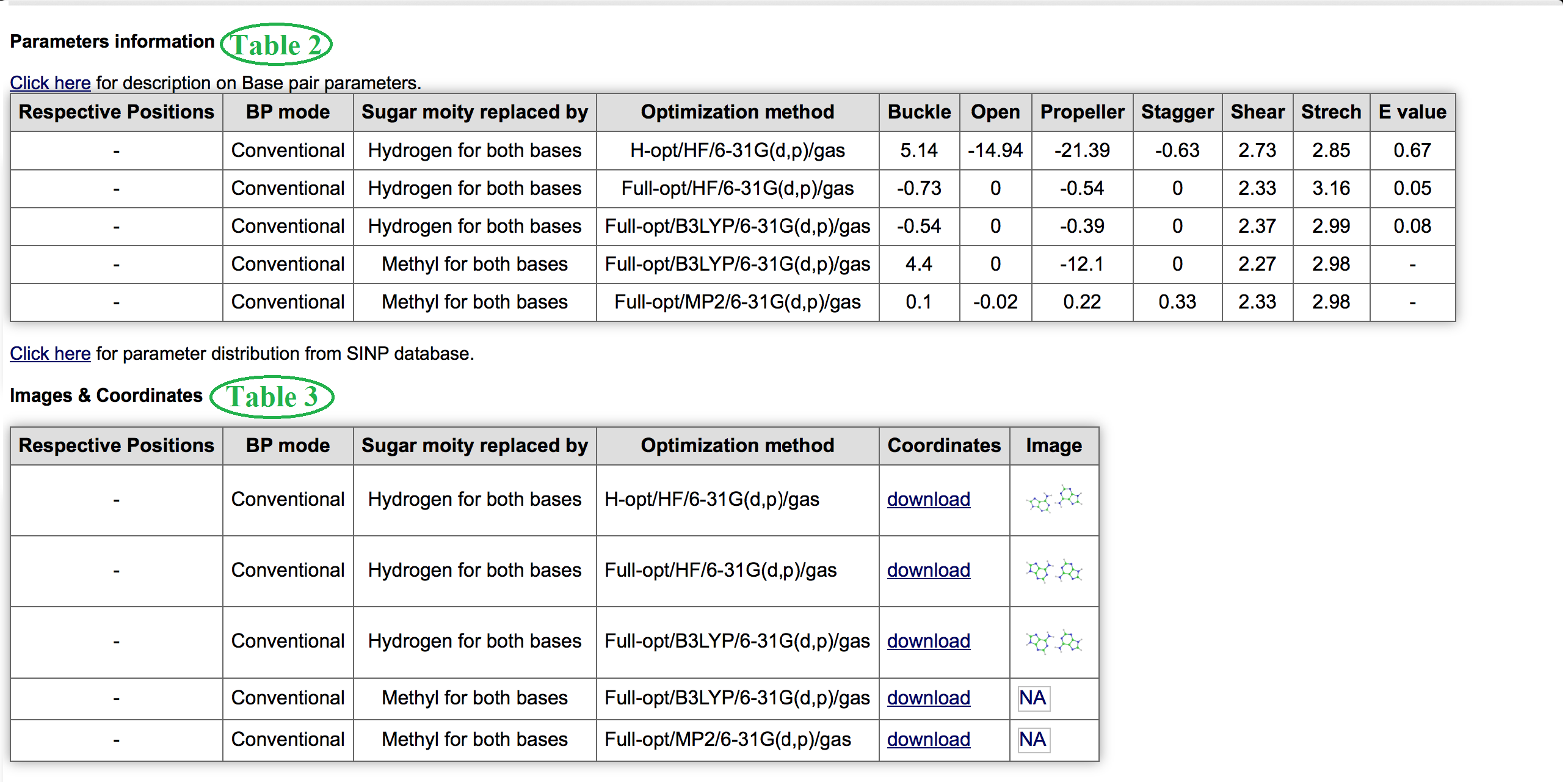
Optimization method mentioned here consists of information about optimization type, method used, basis set used, and phase separated by "/". Click Here to understand each term.
Table 4 and Table 5 marked in Figure 7 provides differences between different optimized geometry in terms of root mean square deviation (RMSD) and measurement of isostericity related parameters respectively. In Table 4 coordinates and images of two superimposed base pairs has been provided with a download option.
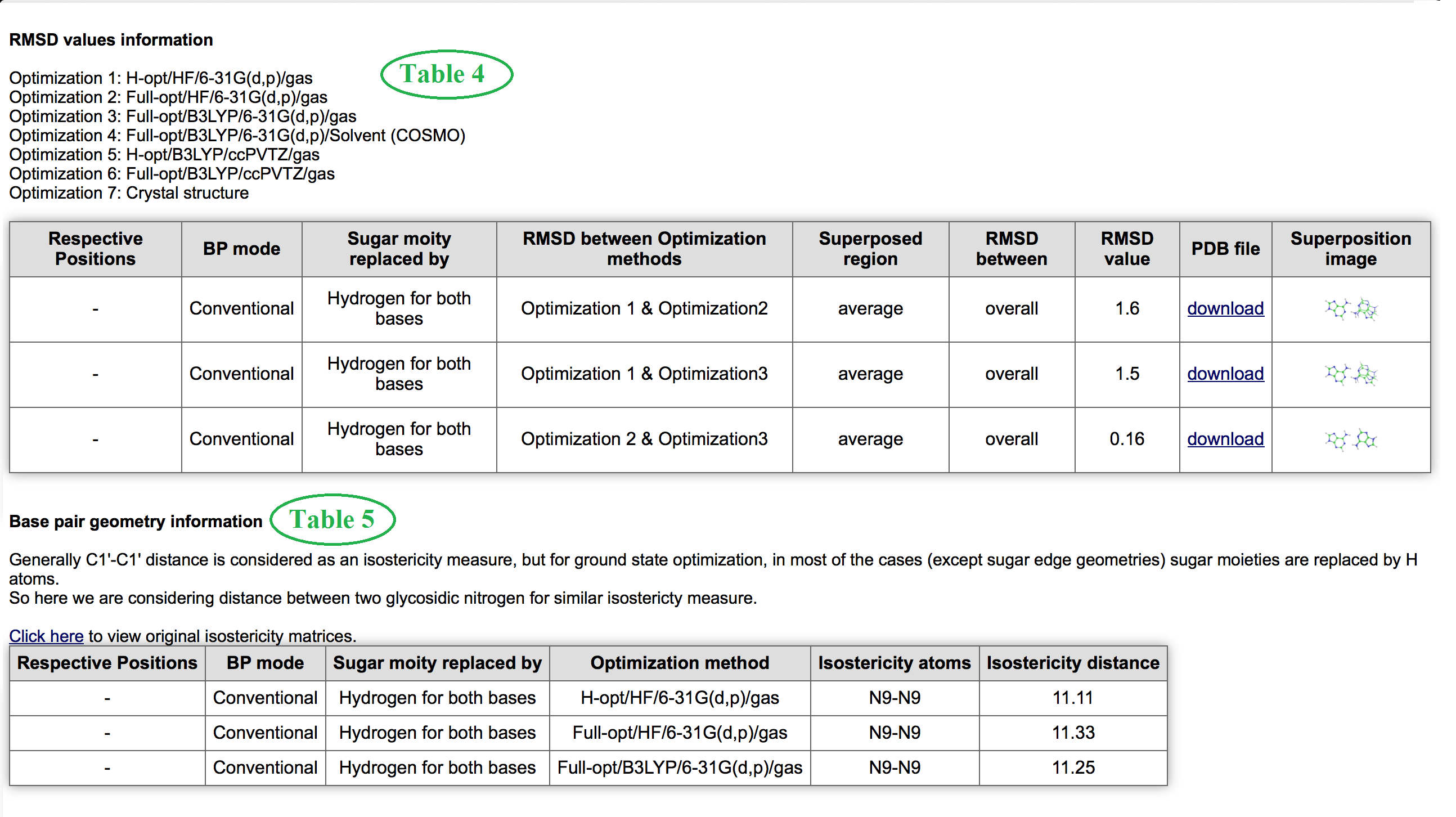
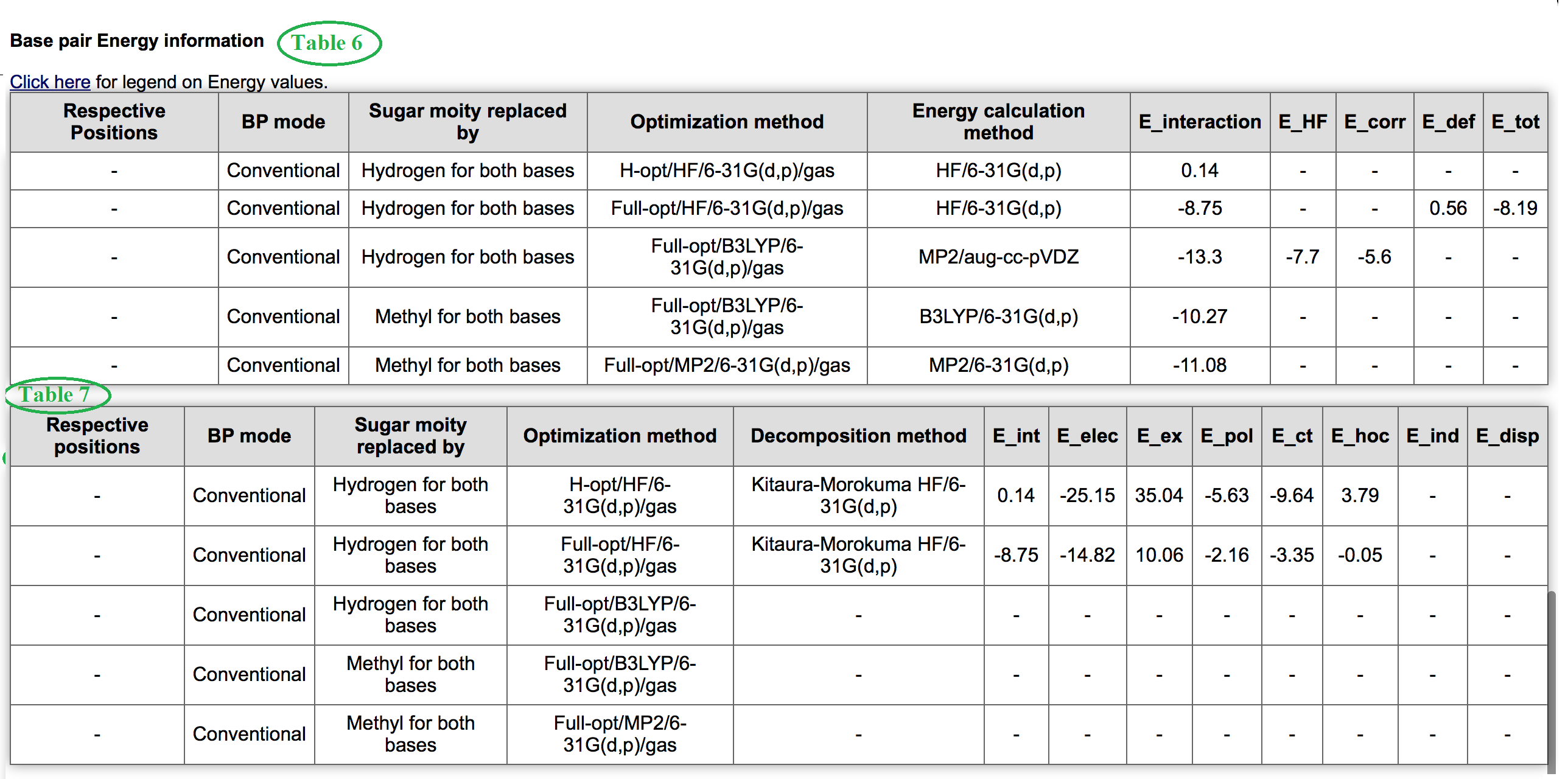
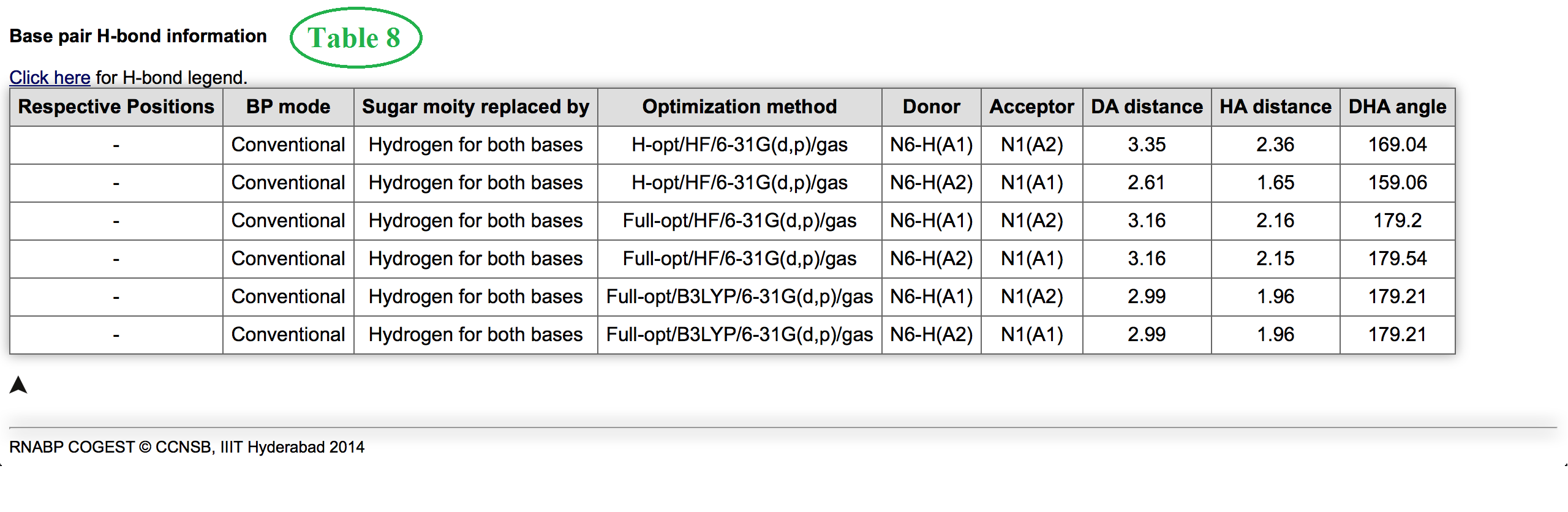
Table 6 and 7 shown in figure 8 consolidate all information related to interaction energy and its components. Know more details about interaction energy calculation methods. Table 7 contains information about energy decomposition analysis. The meaning of each component is defined here. Finally Table 8 marked in Figure 9 contains information about hydrogen bonding pattern, in terms of donor, acceptor atoms identity, donor-acceptor (DA distance) distance, hydrogen-acceptor distance (HA distance and donor-hydrogen-acceptor angle (DHA angle) for a basepair geometry optimized in different methods. These are all about detailed Base Pair Information page.
Go to the home page again and then click on the SEARCH button marked by the green identifier 2. It will redirect you to here, where a number of customized search options are available (marked with green coloured numeric identifiers in figure 10). The functionalities of each of them are given below.
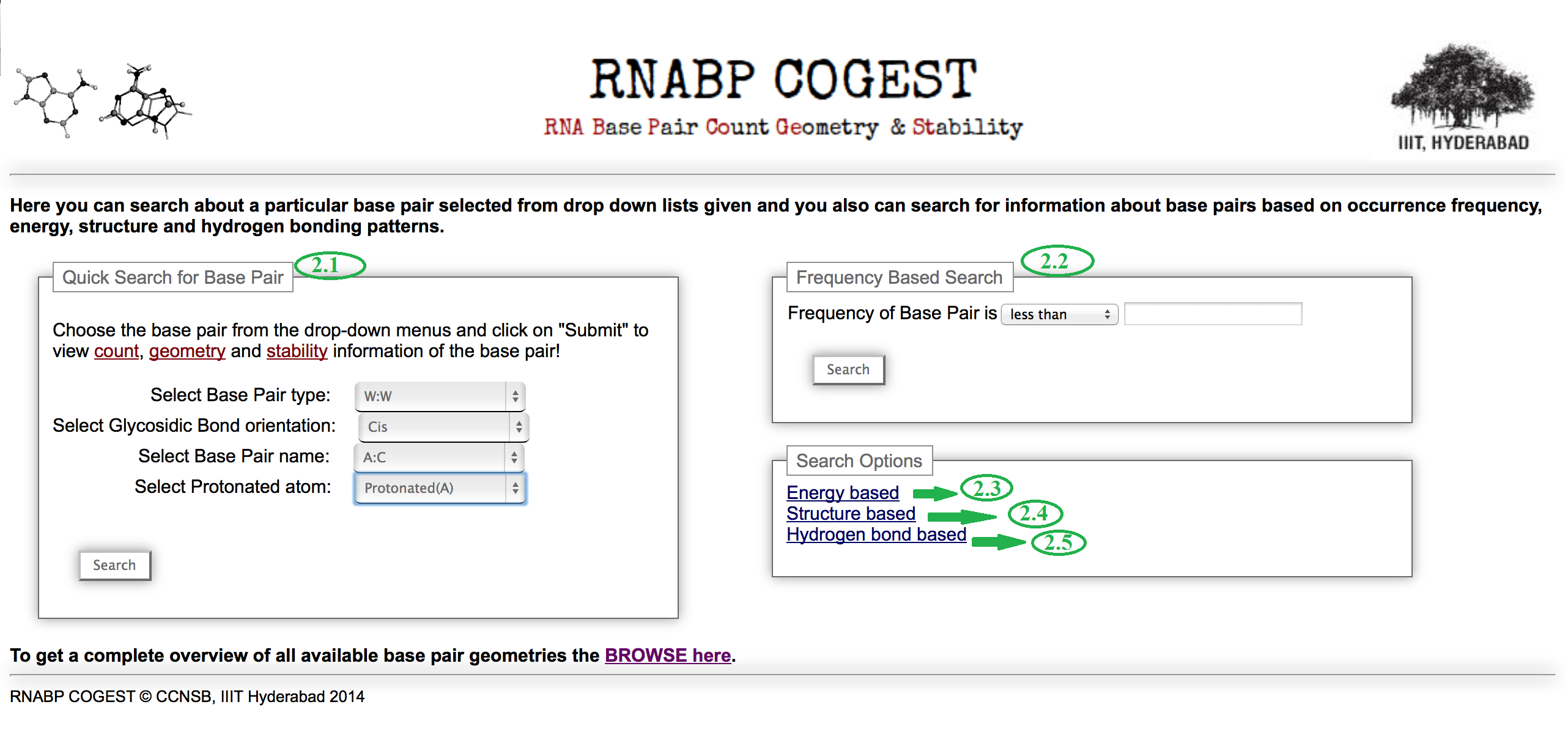
Identifier 2.1: This is a quick search option for retrieving detailed information about a particular base pair type. Instead of searching through the matrix table in browse option if you have any particular base pair in your mind, you can directly choose from the drop down lists given in the box and make a search for it.
It will redirect you to a detailed information page.
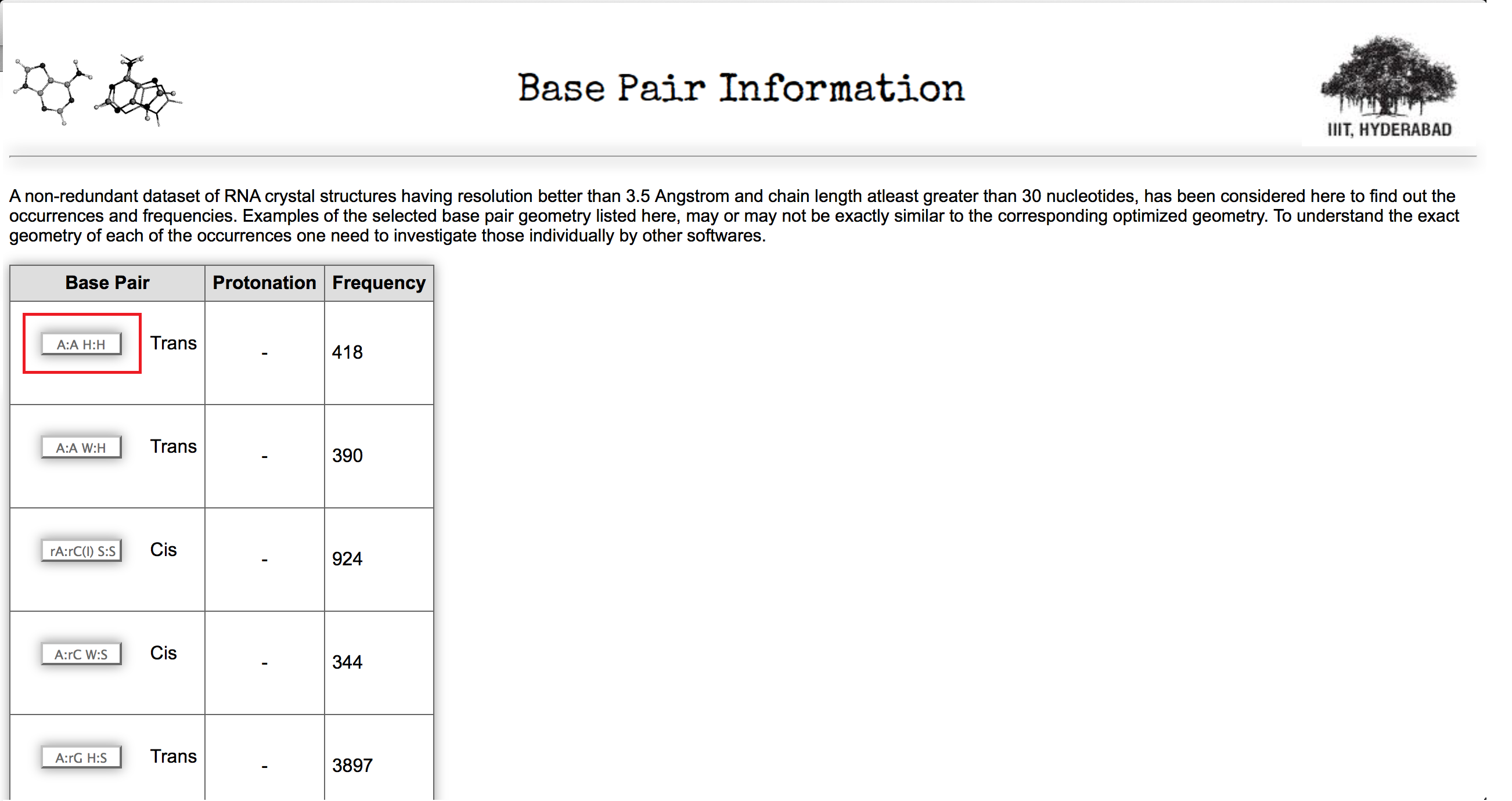
Identifier 2.2: This is a frequency based search option. If you want to search base pairs having a certain range of occurrence frequencies, define the range from the dropdown list and mention the numeric value for the frequency in the text box. It will return you a list of base pairs and their corresponding frequency values. For example, we have made a search for base pairs having occurrence frequency greater than 300. The output is shown in Figure 11. Clicking onto the base pair name (highlighted in red rectangle) it will redirect you to the base pair information page.
Identifier 2.3:
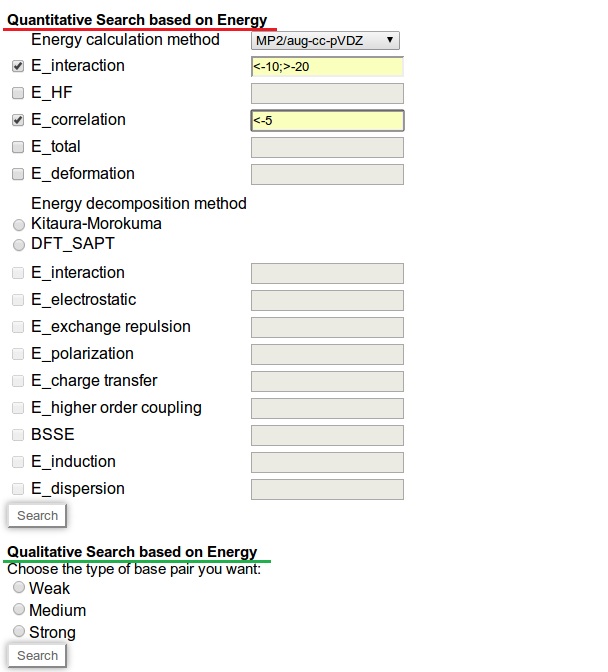
This is a search option based on energy parameters. This search will help you to compare stability of different base pairs.
RNABP COGEST contains intrinsic interaction energy data for all available or modeled non canonical base pairs, calculated by using different QM methods. As interaction energy data have been mostly collected from existing literatures, interaction energy values for all base pairs, reported in the database are not calculated by using same QM method and again one particular QM method has not been implemented for calculating interaction energy of all the base pair geometries. For example, interaction energies of W:W and H:H base pairs have been calculated using MP2/aug-cc-PVDZ method and RI-MP2/aug-cc-PVDZ method is used while calculating interaction energies of sugar edge base pairs. Base pair families and corresponding resource journal artical links are available here. Most of the QM calculation data regarding optimized geometries and interaction energies have been collected from these articles.
Again, for a particular base pair, results can vary depending on the method used. So, it is better to specify energy calculation method from the drop down list before searching with different energy values to compare stability of different base pairs.
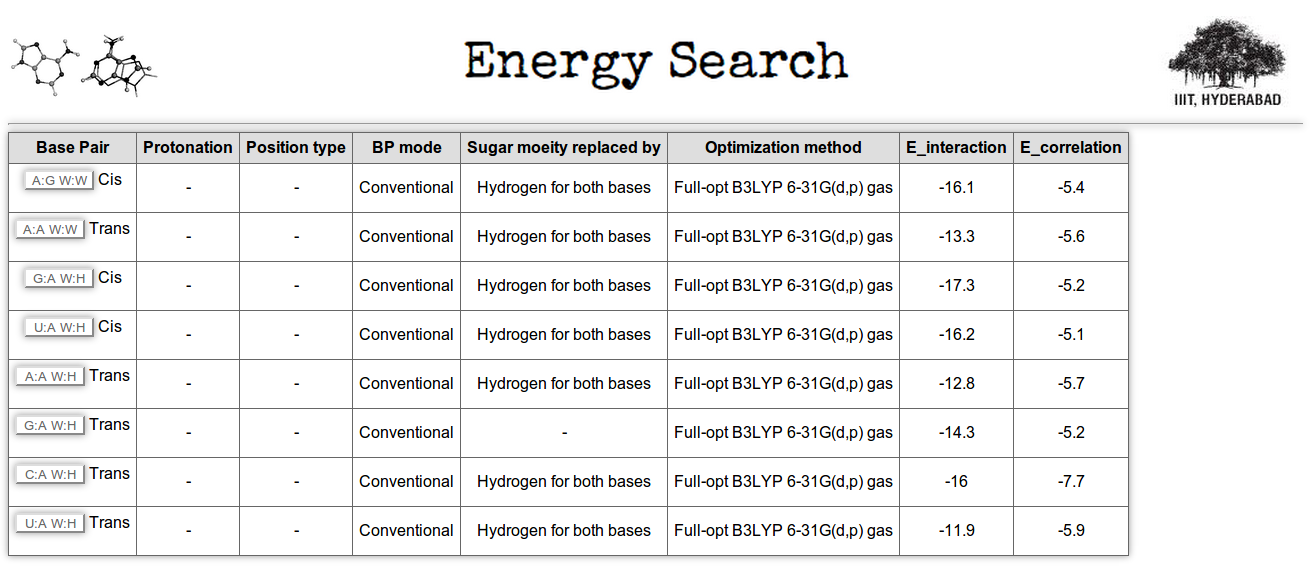
Figure 12 shows the screen shot of the 'Energy search' page. Two types of search options are available here, quantitative (highlighted with red underling in figure) and qualitative (highlighted with green underling in figure). In quantitative search option you can make complex query by selecting multiple parameters and defining the desired range of each parameter. Here, for example in Figure 12, we have made a search for those base pairs having interaction energy within the range of -10 to -20 kcal/mol (units are not required to mention in text box), and the correlation component of interaction energy has more negative value than -5kcal/mol. In qualitative search option you can search for weak, medium or strong base pairs, interaction energy ranges of each type is pre defined as per the existing literature. Figure 13 shows the sample result for the query made in Figure 12.
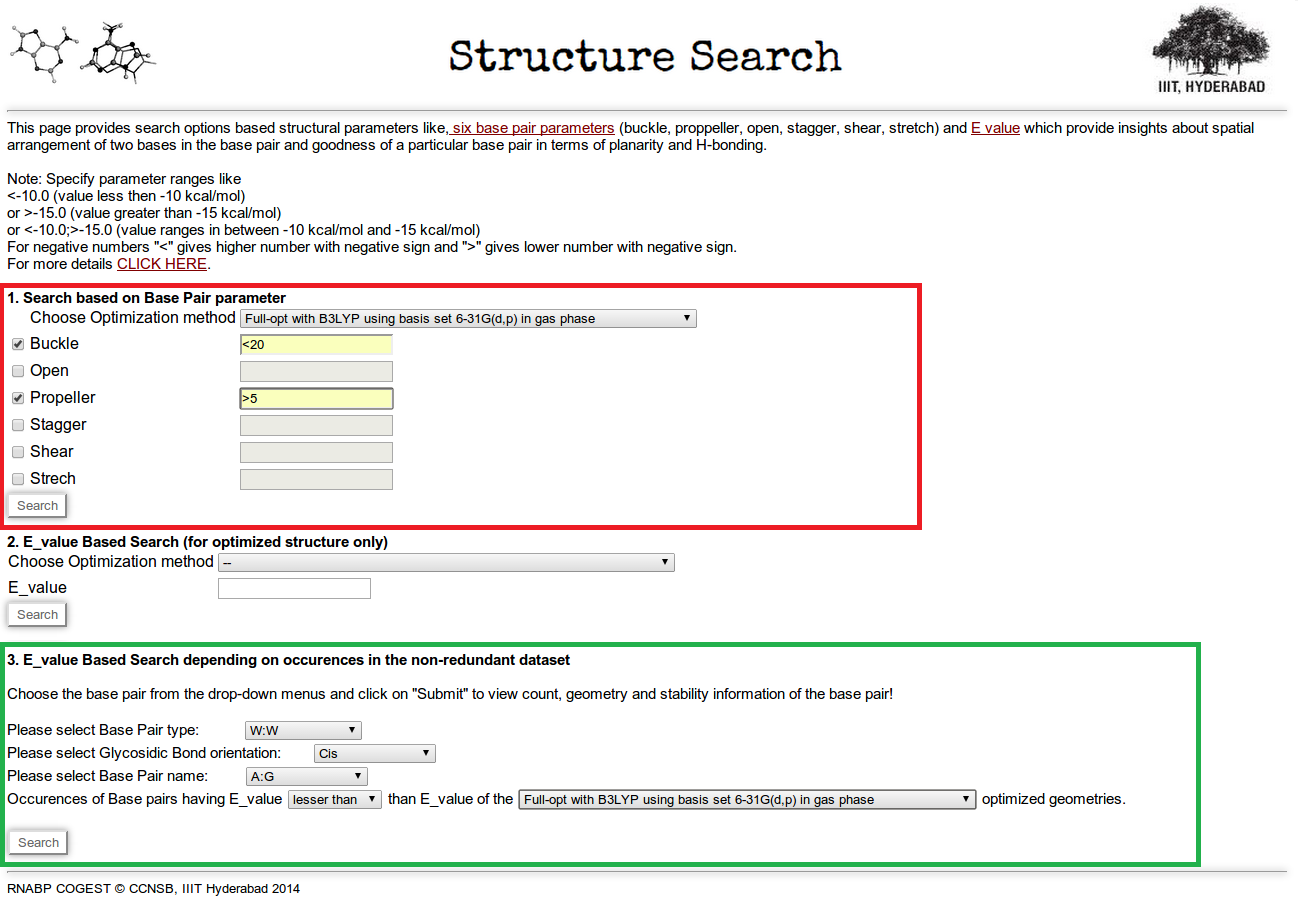
Identifier 2.4: This is a search option based on structural parameters (base pair parameters) and E value. Figure 14 shows the screen shot of structure search page. Here three types of searching options are available.
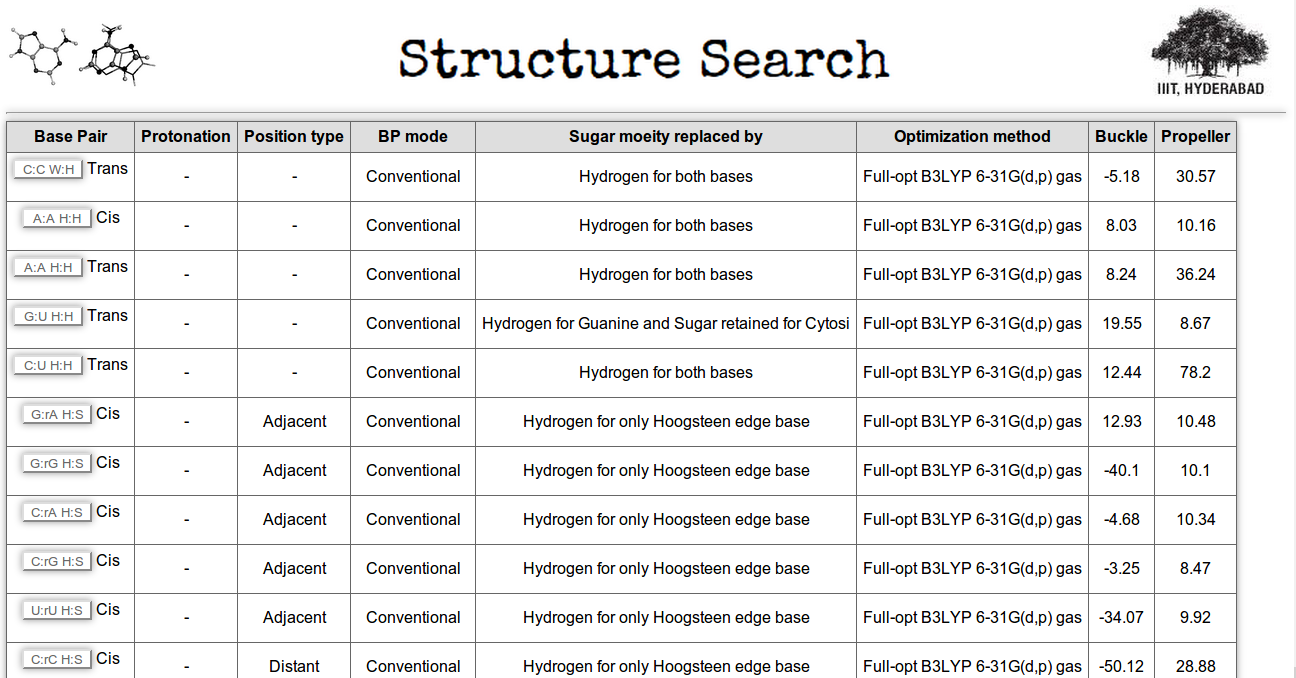
Through the first searching option it is possible to retrieve base pair information having particular range of base pair parameter values. It is also possible to customize the search option by defining specific ranges of multiple base pair parameters at a time. Here users have an option for selecting optimization method, which will give base pair parameters of different ground state optimized structures, where the optimization method is same for all. In Figure 14 (highlighted in red box) we have made a search for base pairs having buckle less than 20.0 degree and propeller greater than 5.0 degree. Result is shown in Figure 15.
Through the second searching option you can search for hydrogen optimized and full optimized base pairs that lie under specified E_value ranges.
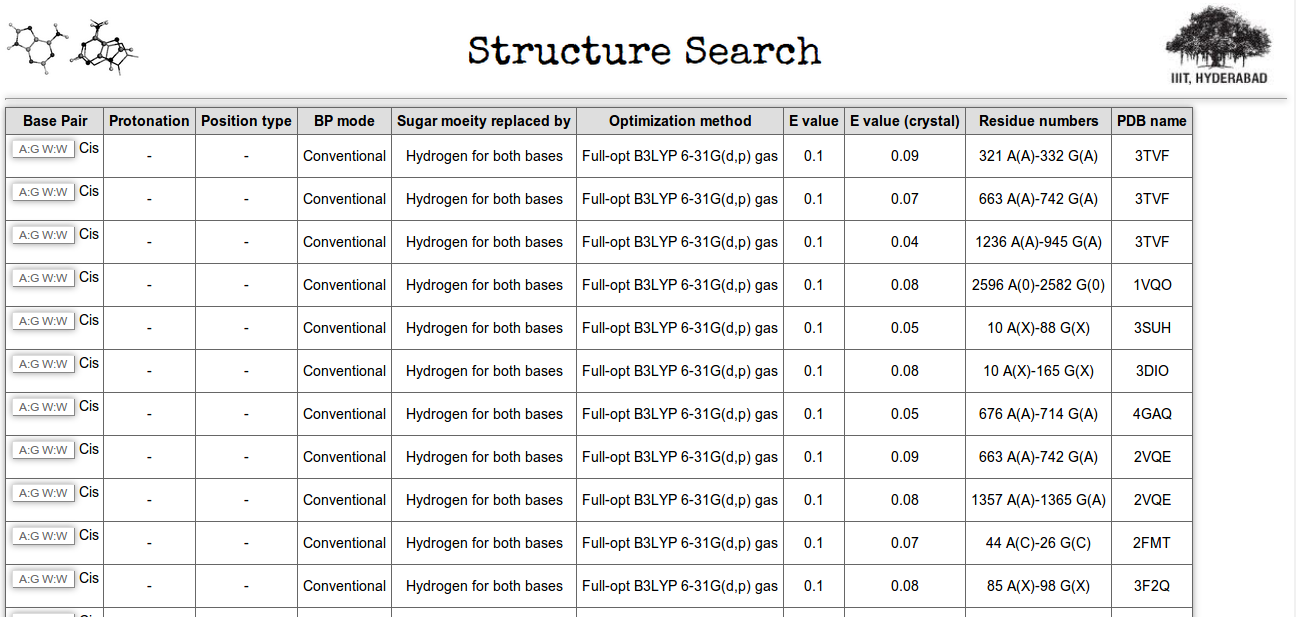
Through the third searching option user can obtain occurrences of a particular base pair in the non redundant crystal structure dataset, having E value better or poorer than its corresponding full optimized structure. In Figure 14 (highlighted in green box) we have made a search for all the occurrences of A:G W:W Cis base pairs having lesser E_value (better geometry) than its corresponding optimized structure. Figure 16 shows the result.
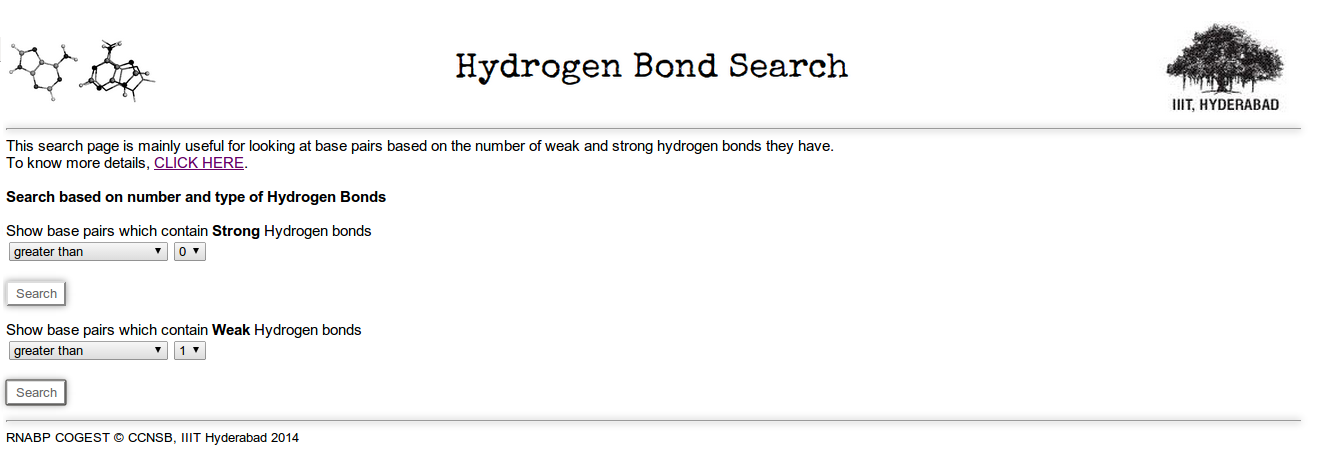
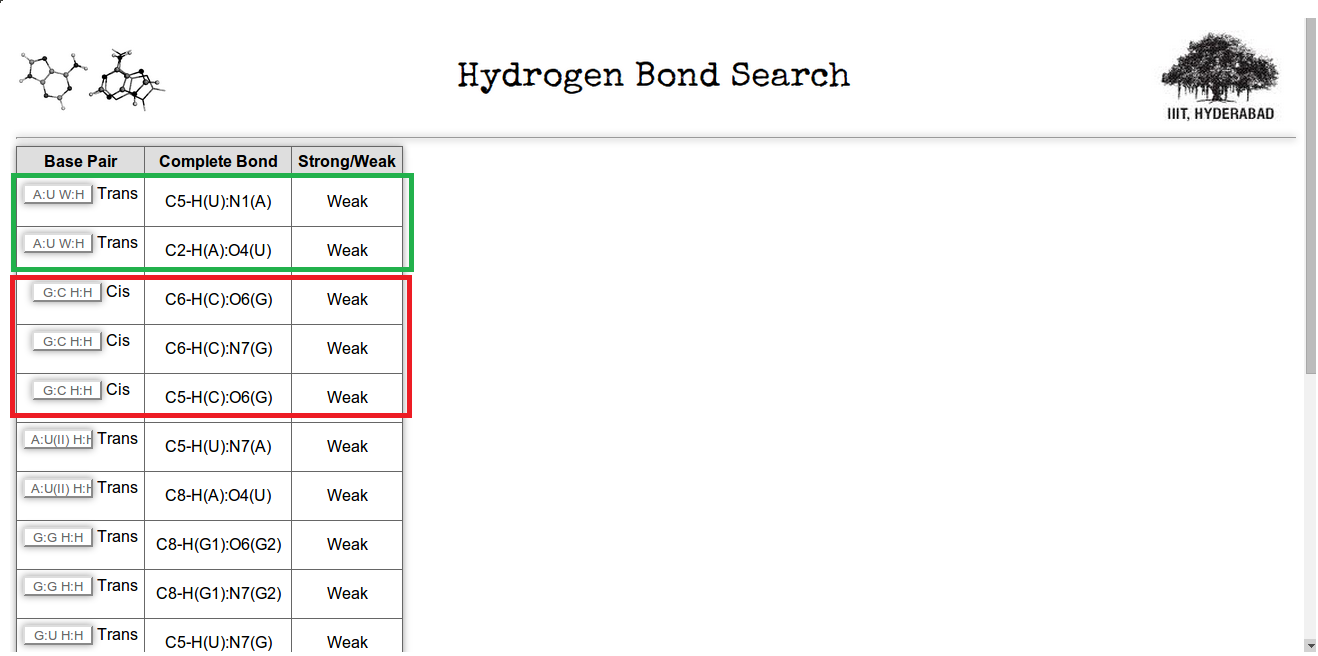
Identifier 2.5: Here you can search for base pairs by mentioning numbers of strong or weak Hydrogen bonds (link to legend page). Figure 17 shows the screen shot of h-bond based search page. In the figure we have made a search for base pairs having more than 1 weak hydrogen bonds (C-H..O, C-H..N).
The result of the previous search is displayed in Figure 18. The search result contains all the base pairs having more than one week hydrogen bonds with its complete hydrogen bonding details. For example in figure 18 A:U W:H Trans(highlighted in green box) has two and G:C H:HCis (highlighted in red box)has three week hydrogen bonds, to stabilize the structures.
Identifier 3: Terms and definition page consists of brief scientific details and important links for most of the area covered in the database. It will help you to understand the data even in a better way. Identifier 4: These are other important database links which contains information about non canonical base pairs. Identifier 5: Some important guidelines to understand the database "Non-canonical RNA Base Pair Database", linked with RNABP COGEST as a companion databas, developed and maintained by our collaborators at Saha Institute of nuclear physics, Kolkata. Basically from here you will get a pdf file where notaions and abbreviations used in both the databases are listed. Identifier 6: Here users can give their feed back by mentioninig their details. Your feed back will definitely help us for further development of RNABP COGEST.