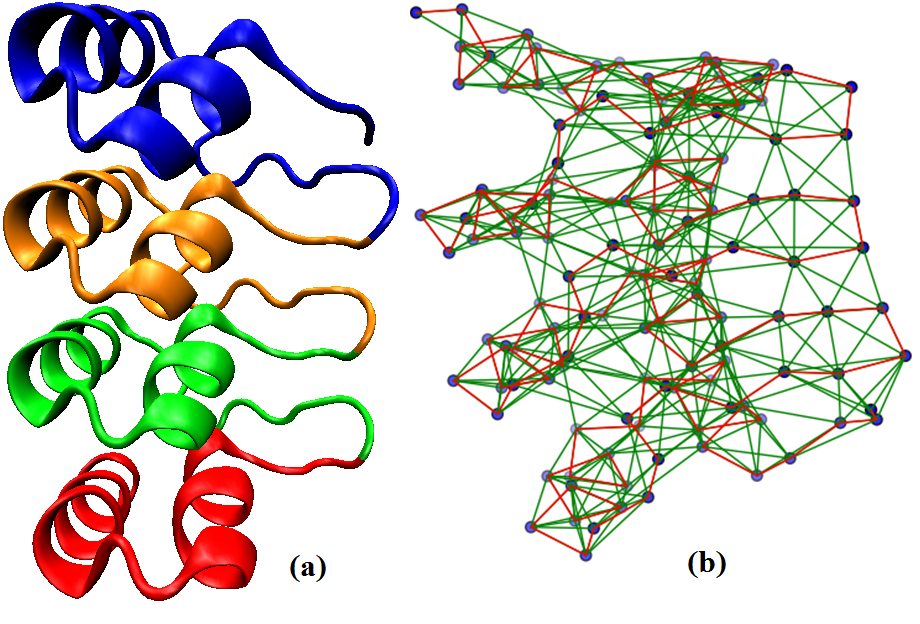
Structure based identification of Ankyrin repeats by graph spectral approach
A Protein Contact Network (PCN) is constructed from a 3D protein structure
as input by considering Cα atoms of amino acids as nodes and an edge drawn
between them if the distance between the Cα atoms is below a predefined
threshold (~7Å), capturing backbone and non-covalent interactions. This
connectivity information is represented in the adjacency matrix. The
repeat copies are predicted based on the conserved pattern in the eigenvector
profile corresponding to the largest eigen value of adjacency matrix
(eigenvector centrality) and secondary structure information from STRIDE database.
|
|
|
|
|
|
|
Related publication: Chakrabarty B, Parekh N: Identifying tandem Ankyrin repeats in protein structures. BMC Bioinformatics 2014, 15:6599. |
|
![]()
Contact: Broto Chakrabarty and Dr. Nita Parekh,
Centre for Computational Natural Sciences and Bioinformatics, International Institute of Information Technology-Hyderabad
email: broto.chakrabarty@research.iiit.ac.in, nita@iiit.ac.in